Drug Metabolism Evaluation Technical Services
I. Model Overview
Drug metabolism refers to the biochemical transformation of drugs within an organism, ultimately converting them into metabolites for excretion. It is a critical process governing drug efficacy, toxicity, and elimination. While the liver is the primary site of drug metabolism, other tissues (e.g., intestine, kidney, lungs) may also contribute. Metabolism modifies drugs into more excretable forms while altering their pharmacological activity or toxicity. Key metabolic sites include:
Liver: Rich in metabolic enzymes (e.g., CYP450, UGTs).
Intestine: Involves gut microbiota and enterocytes.
Kidney/Lungs: Participate in specific metabolic pathways.
1. Metabolic Pathways of Small-Molecule Compounds
Small-molecule drugs primarily undergo Phase I and Phase II metabolism:
2. Metabolic Pathways of Small Nucleic Acid Drugs
Small nucleic acid drugs (e.g., siRNA, ASO) are metabolized via distinct mechanisms:
II. Detection Models
1. Primary Hepatocytes
Species: Human, monkey, dog, rat, mouse.
Applications:
2. S9 Fraction & Liver Microsomes
Liver Microsomes:
Phase I metabolism studies (oxidation, reduction, hydrolysis).
Metabolite identification via LC-MS/MS.
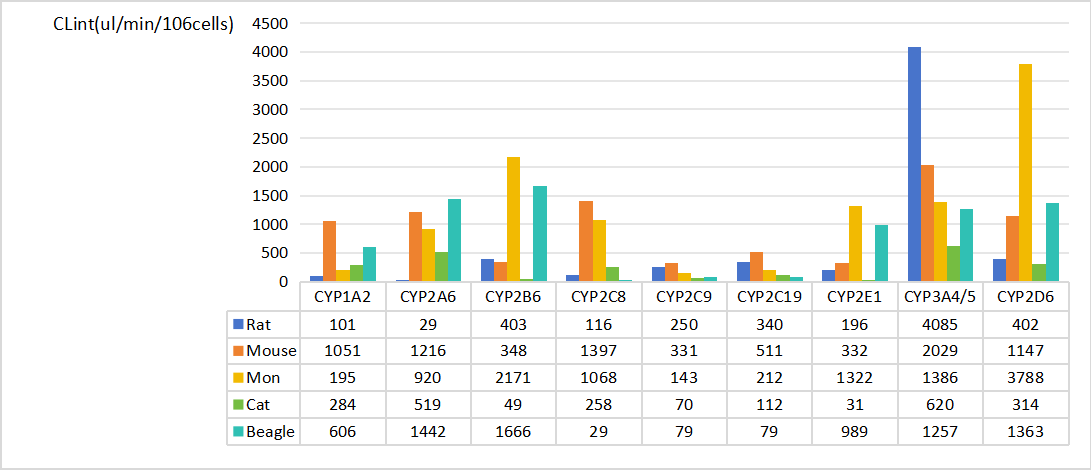
Enzyme phenotyping and kinetic studies (t1/2, Clint).
Derived from endoplasmic reticulum membranes, rich in Phase I enzymes (CYP450) and limited Phase II enzymes (UGTs, STs).
Applications:
Liver S9 Fraction:
Complex metabolite profiling (non-CYP pathways).
Enzyme activity assessment (lower activity but broader enzyme coverage).
Contains both Phase I and Phase II enzymes.
Applications:
Key Differences:
Microsomes: CYP-dependent metabolism.
S9: Non-CYP or multi-phase metabolism.
Enzyme Activity: Microsomes (higher for Phase I) vs. S9 (lower but comprehensive).
Use Cases:
3. Lysosomes
Drug Development & Targeted Therapy:
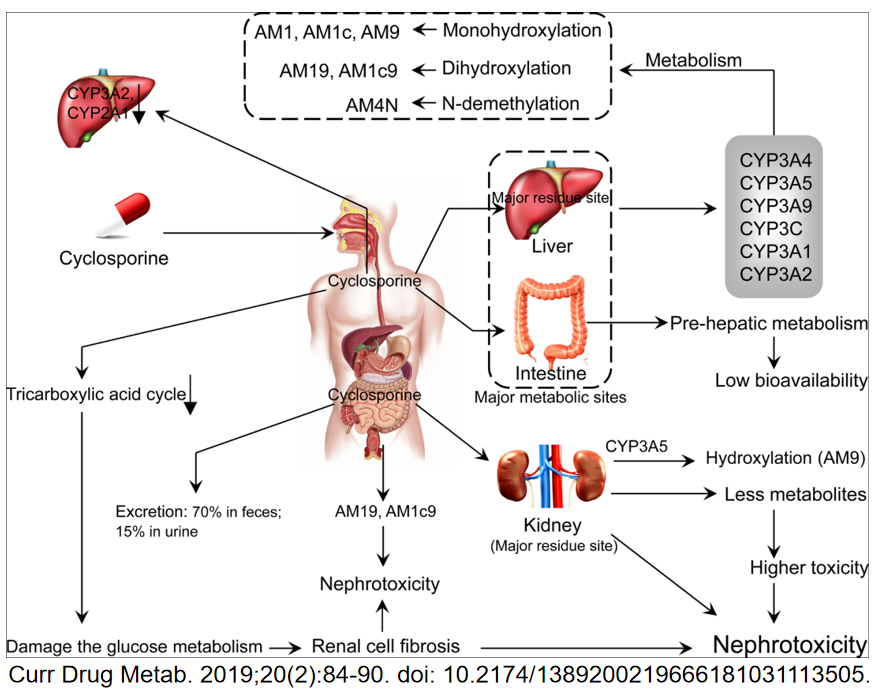
Lysosome-Targeted Drugs: Exploit acidic environments/enzymes (e.g., Polyphyllin D targeting SMPD1 to induce lysosomal membrane permeabilization [LMP] in HCC).
Overcoming Drug Resistance: Disrupt lysosomal drug sequestration (e.g., reversing sorafenib resistance).
Protein Degradation Technologies:
Metabolic Research & Disease Models:
Study lysosomal enzyme activity (e.g., density gradient ultracentrifugation, mass spectrometry).
Model diseases linked to lysosomal dysfunction (e.g., neurodegenerative disorders, cancer).
Drug Metabolism & Stability:
Autophagy & Cell Death Regulation:
III. Detection Conditions

LC-MS/MS System
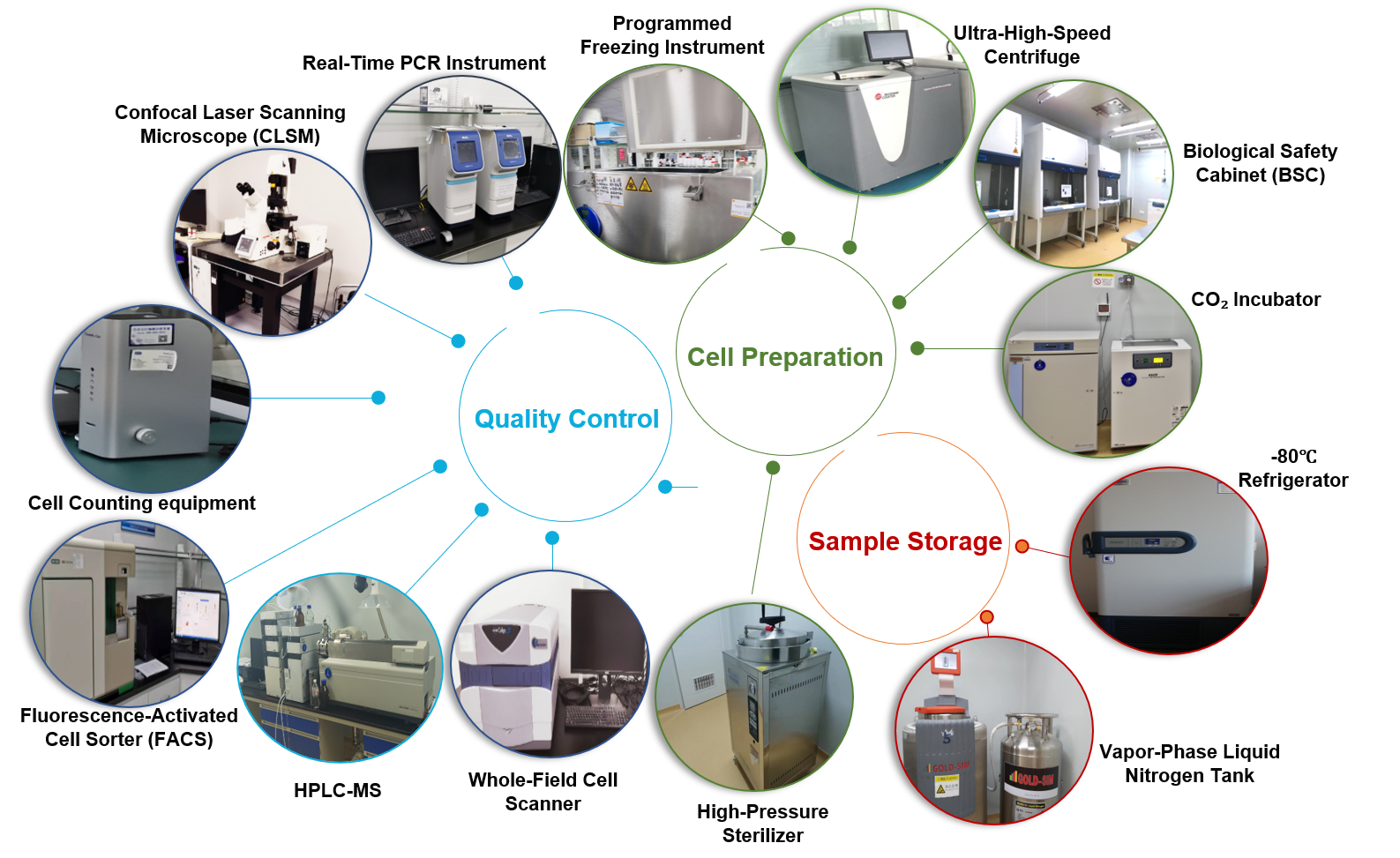
IV. Service Credentials, Facilities, and Team

Expert Team:
Ph.D., Wuhan Institute of Virology, CAS.
14+ years in liver research.
Principal investigator for 7 national/provincial grants (NSFC, Guangdong DST, Shenzhen STIC).
10+ first/corresponding-author SCI papers.
Led by AIMBE Fellow, National Young Thousand Talents, and Shenzhen High-Level Talent Awardees.
Dr. Ming Zhou
V. Case Studies
Case 1: A Shanghai-Based Biopharma Company
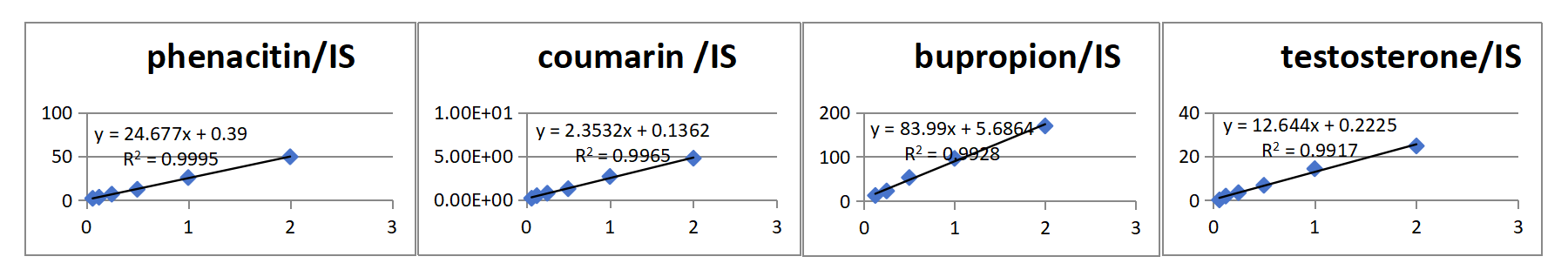
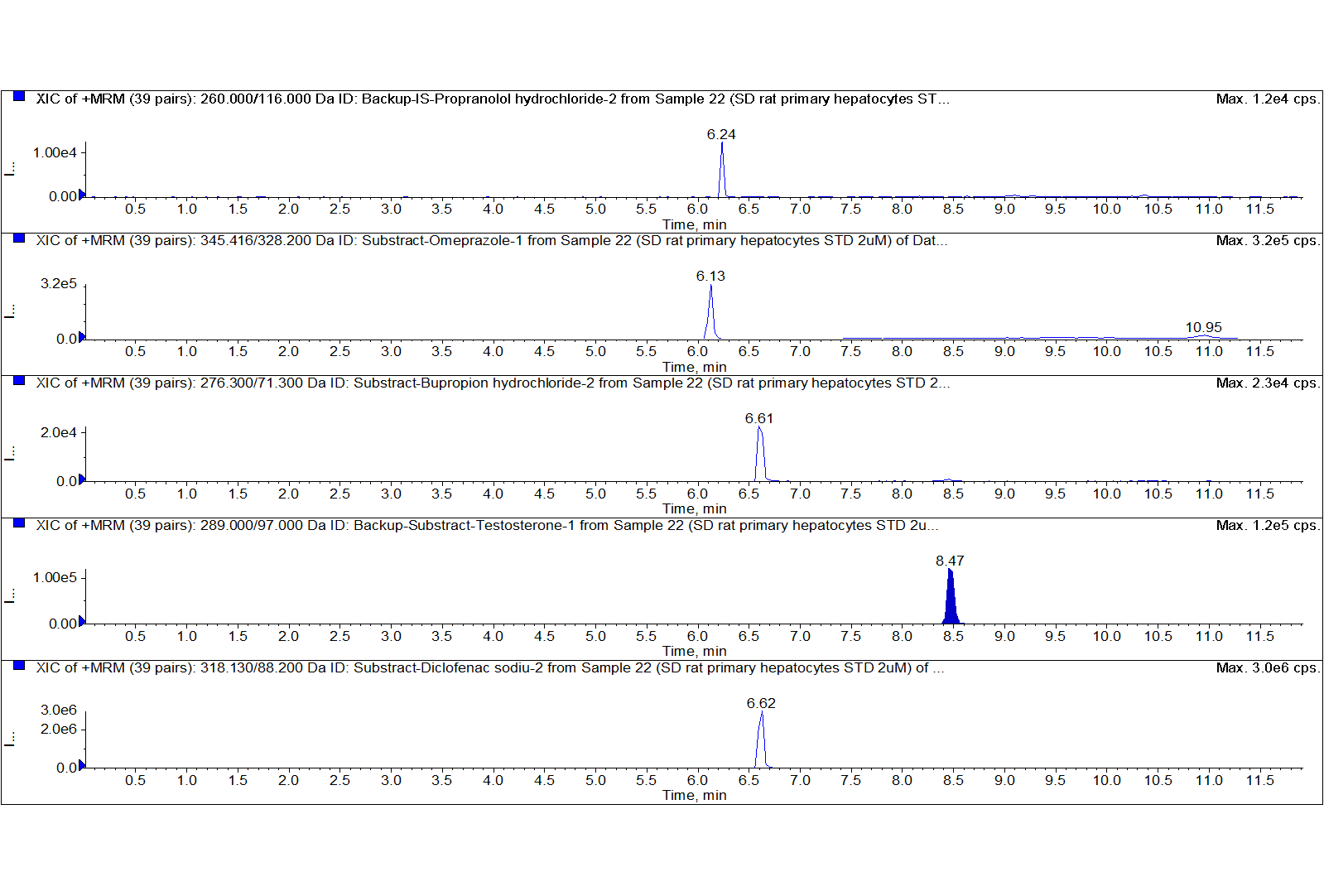
Case 2: A Hangzhou Research Institute